Date: Wed, 21 Jan 2015 09:31:16 +0530
Hi Dan
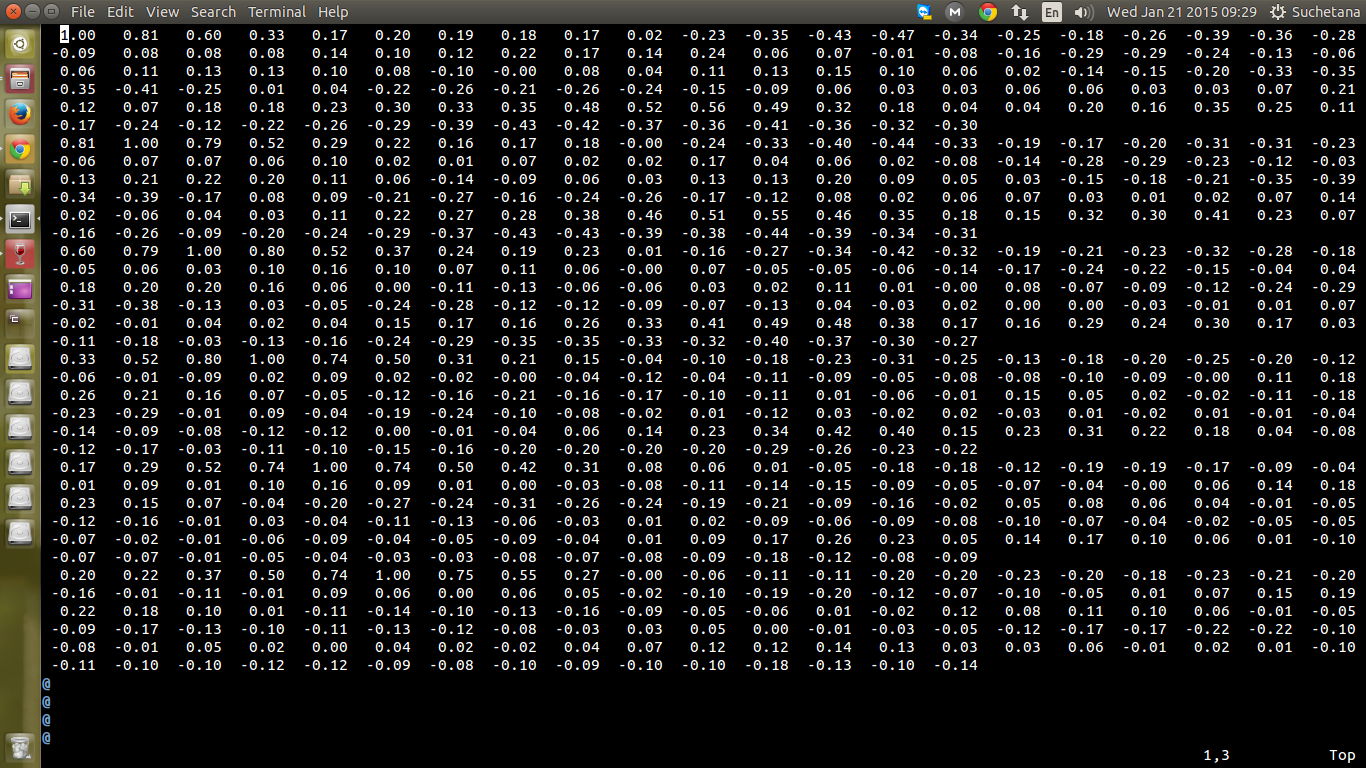
Thanks for the suggestions. I tried using small residue selections and they
are working fine in Amber12. But the moment I increase the number of
residues, the output file doesn't contain the data in the correct matrix
format. I am sure I am getting 120x120 values, but the output is not in a
matrix format. I am attaching a screenshot if it helps.
Any help would be appreciated.
Thanks
On Tue, Jan 20, 2015 at 11:49 PM, Daniel Roe <daniel.r.roe.gmail.com> wrote:
> Hi,
>
> On Tue, Jan 20, 2015 at 6:40 AM, Suchetana Gupta bt13d072
> <bt13d072.smail.iitm.ac.in> wrote:
> > trajin last_20_stripped_imaged.cdf
> > rms first mass :1-120
> > matrix correl :1-120.CA out last_20_wt_correl.txt byres
> >
> > However, the output matrix file is not in a 120 X 120 format. Instead, I
> > see data in a random fashion. So much so, that I am unable to understand
> > which value corresponds to which residue. Is there any method to get the
> > output in a 120X120 matrix?
>
> It's not clear what you mean by "random fashion". Have you run the
> tests for your Amber installation? You should absolutely get a file
> containing 120 rows and 120 columns. Try it on a smaller number of
> residues first to become familiar with the output. For example, a 4
> residue selection (:1-4) will give output containing 4 rows and 4
> columns like so:
>
> 1.00 0.38 0.23 0.02
> 0.38 1.00 0.46 -0.40
> 0.23 0.46 1.00 0.24
> 0.02 -0.40 0.24 1.00
>
> The first row and column is residue 1 to 1, the first row and second
> column is residue 1 to 2, etc. Hope this helps,
>
> -Dan
>
> PS - Also, consider upgrading to AmberTools 14 (which is free), as it
> contains more functionality and bug fixes. Also consider using cpptraj
> instead (ptraj is no longer being actively developed).
>
> >
> > Thanks in advance for your help
> > Suchetana Gupta
> > IIT Madras
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org
> > http://lists.ambermd.org/mailman/listinfo/amber
>
>
>
> --
> -------------------------
> Daniel R. Roe, PhD
> Department of Medicinal Chemistry
> University of Utah
> 30 South 2000 East, Room 307
> Salt Lake City, UT 84112-5820
> http://home.chpc.utah.edu/~cheatham/
> (801) 587-9652
> (801) 585-6208 (Fax)
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screenshot_from_2015-01-21_09:29:59.png)
