Date: Fri, 16 Jan 2015 11:01:15 +0100
Hello all,
I have been trying to compute the principal axes for two 4mer water
clusters and have ran into what I think is a discrepancy in the
calculations. The structure of both clusters is the same, but they differ
in their input coordinates and in the ordering of the TP3 water residues -
by that I mean if you start with residue 1 and proceed clockwise, the
residue numbering goes 1,3,4,2 (4mer-1 in red) versus 1,2,3,4 (4mer-2 in
blue). (As you might guess these structure were obtained from an MD
simulation, and thus the difference in the water residues ordering.)
Using cpptraj (fully patched AmberTools14), I then compute the principal
axes of each cluster. I expected that I obtain the same results. Instead if
I view the resulting principal axes, overlayed with the output cluster
structures (as obtained from the dorotation keyword), I see that the axes
are indeed identical, but the clusters are offset. I have also tried the
"origin" keyword (both before and after the principal command) to help
correct the structures' offset, but this doesn't change the final
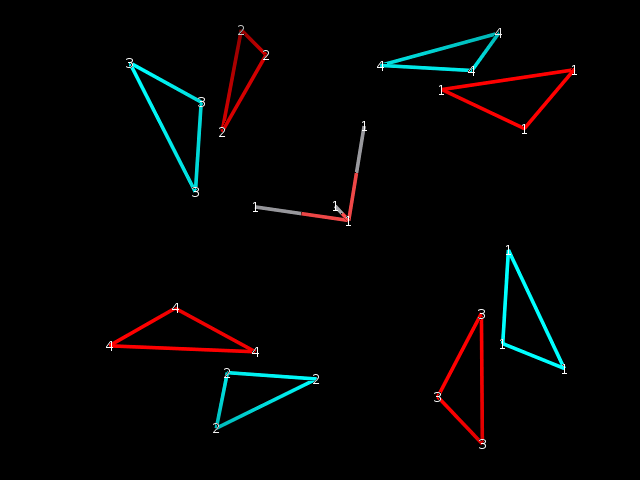
coordinates within the mol2 file. As seen in the image, the 4mer-2 has two
of its principal axes pointing directly at hydrogen atoms, while the 4mer-1
has the same two axes pointing to the center of hydrogen bonds.
I would appreciate if someone could point out if I have made an error in
my input or in my thinking. My only guess(!) is that perhaps something is
going wrong with dorotation within the principal command.
Just to be clear, the attached image shows the output structures (eg.
4mer-1.mol2) and principal axes (e.g. 4mer-1.x.pdb) as obtained from
cpptraj, whose input for the 4mer-1 is below. The two clusters would
overlay perfectly if one did it manually within a molecular viewer - by an
~90 degree rotation of the blue cluster.
cpptraj.in:
trajin 4mer-1.leap.pdb
center :1-4 origin
principal :1-4 dorotation mass
trajout 4mer-1.mol2
vector v1 :1-4 principal x out 4mer-1.x.dat
vector v2 :1-4 principal y out 4mer-1.y.dat
vector v3 :1-4 principal z out 4mer-1.z.dat
vector v4 :1-4 principal x trajout 4mer-1.x.pdb trajfmt pdb
vector v5 :1-4 principal y trajout 4mer-1.y.pdb trajfmt pdb
vector v6 :1-4 principal z trajout 4mer-1.z.pdb trajfmt pdb
Thanks in advance,
Karl
-- Karl. N. Kirschner, Ph.D. Research Associate Bonn-Rhein-Sieg University of Applied Sciences Grantham-Allee 20, 54757 Sankt Augustin, Germany
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: output.png)
