Date: Thu, 15 Jan 2015 17:17:18 +0200
Dear Users,
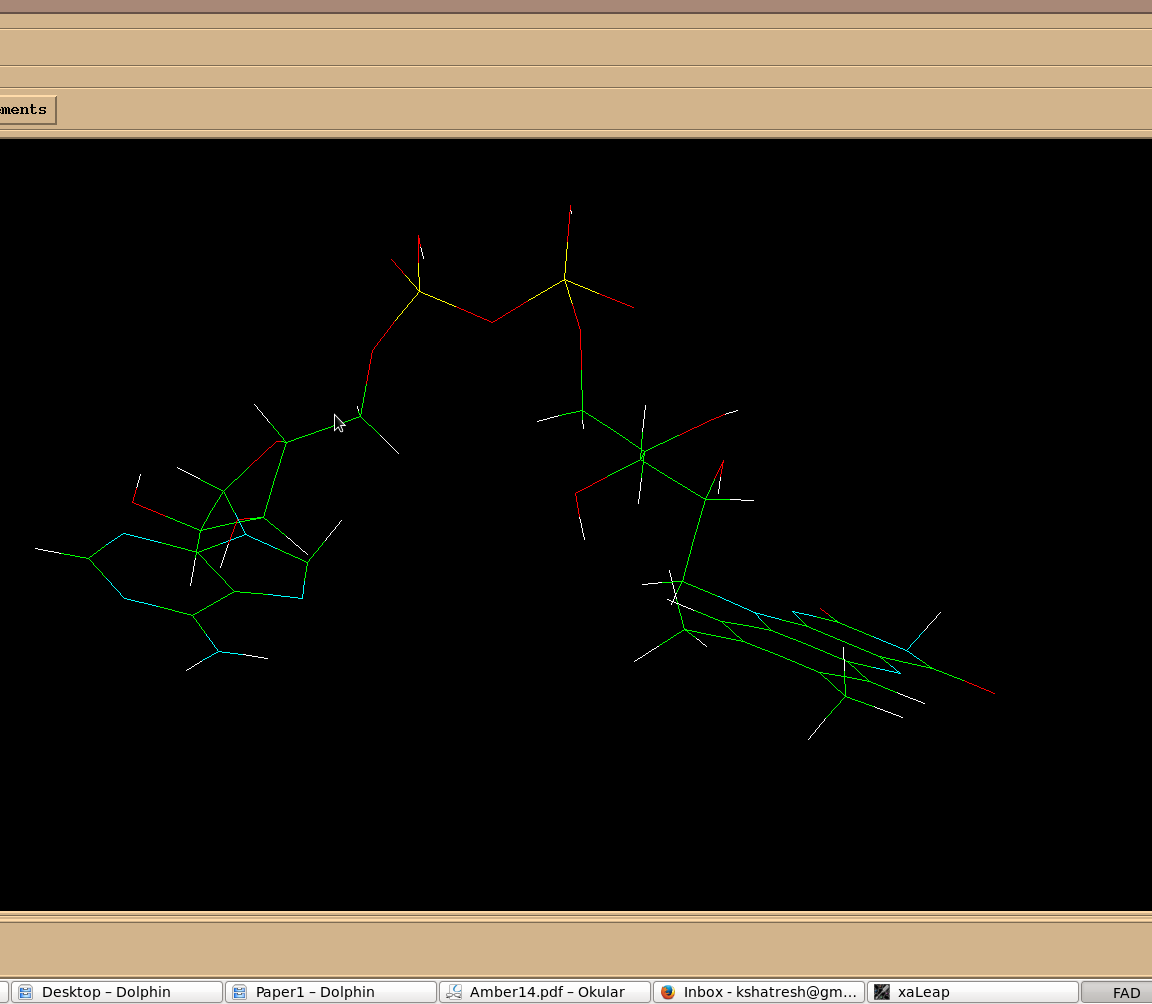
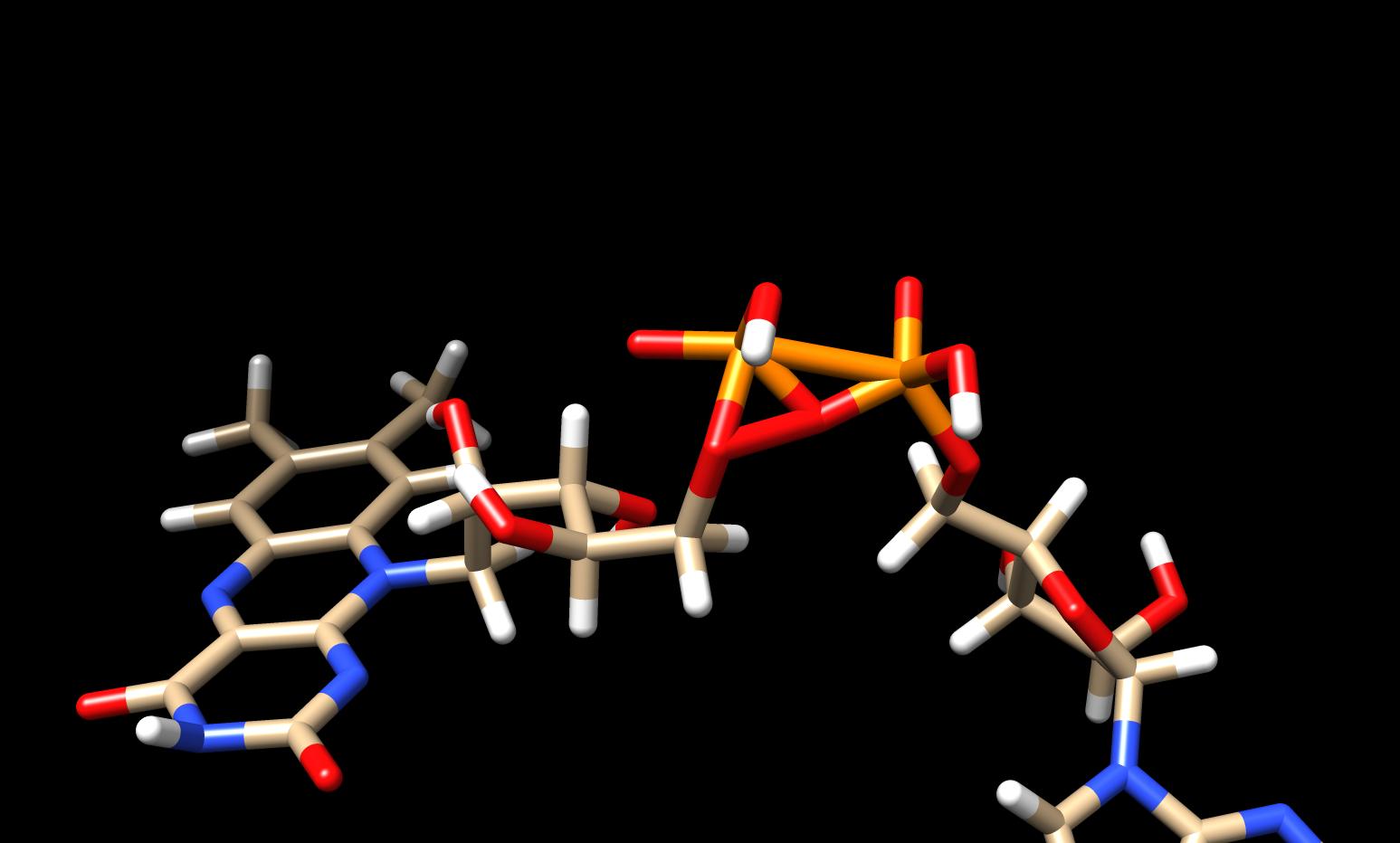
I am simulating flavin adenine dinuecleotide complex. During the
minimisation, the atoms neighbouring to two phosphorus of FAD, stack
together and are giving 1-4 ELE errors. However the parameterized structure
of FAD looks fine in leap. I have attached two snapshots before and after
minimisation which may clear my issue.
I will be very thankful for every suggestion to fix the problem.
Regards
Kshatresh
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screenshot.png)
(image/jpeg attachment: image.jpeg)
