Date: Tue, 4 Mar 2014 00:29:00 +0800
Dear bill,
Like you I used idecomp=2 but now I am confused regarding the output of "FINAL_DECOMP_MMPBSA.dat". I have attached the snapshots for the following queries:
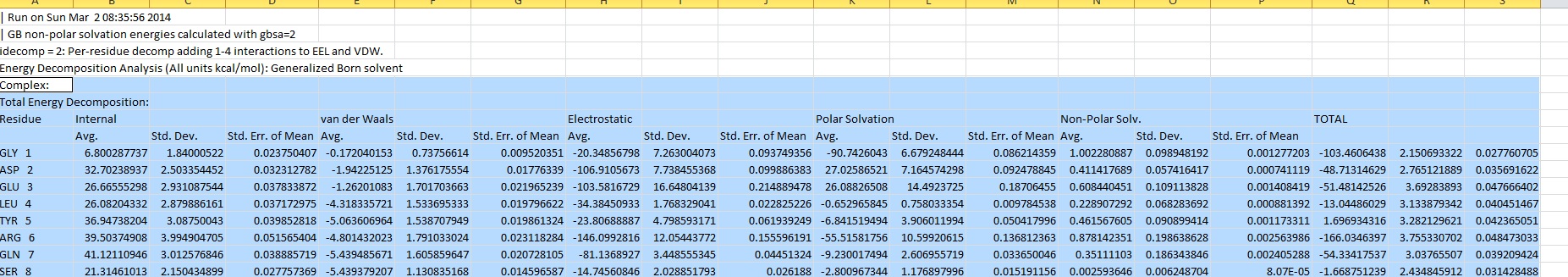
1) As you can see in decomposition_1 which is my output using idecomp=2 is quite different from decomposition_tutorial which is result of idecomp=1 used in amber tutorial http://ambermd.org/tutorials/advanced/tutorial3/py_script/section6.htm . I would like to know what does this section "Total" in decomposition_1 signifies? Unlike other sections like van der waals, electrostatic it has only one sub-section Std. Err. of mean.
Also the difference between outputs I.e. decomposition_1 and decomposition_tutorial is due to different options selected or due to different versions of AMBER. I am using amber12
2) Secondly, what does delta section signifies? As can be seen from decomposition_2 only ligand i.e UNK 155 has significant value
I am really sorry if you find my questions are too na´ve.
Thanks and regards,
Nitin Sharma
Department of Pharmacy,Faculty of Science,National University of Singapore,block S7, Level 2, Singapore : sharmanitin.nus.edu.sg<mailto:sharmanitin.nus.edu.sg> ; http://www.linkedin.com/in/imsharmanitin
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: decompositon_1.jpg)

(image/jpeg attachment: decompositon_2.jpg)

(image/jpeg attachment: decompositon_tutorial.jpg)
