Date: Tue, 25 Feb 2014 19:45:11 +0200
Dear Prof. Walker and Callum Dickson
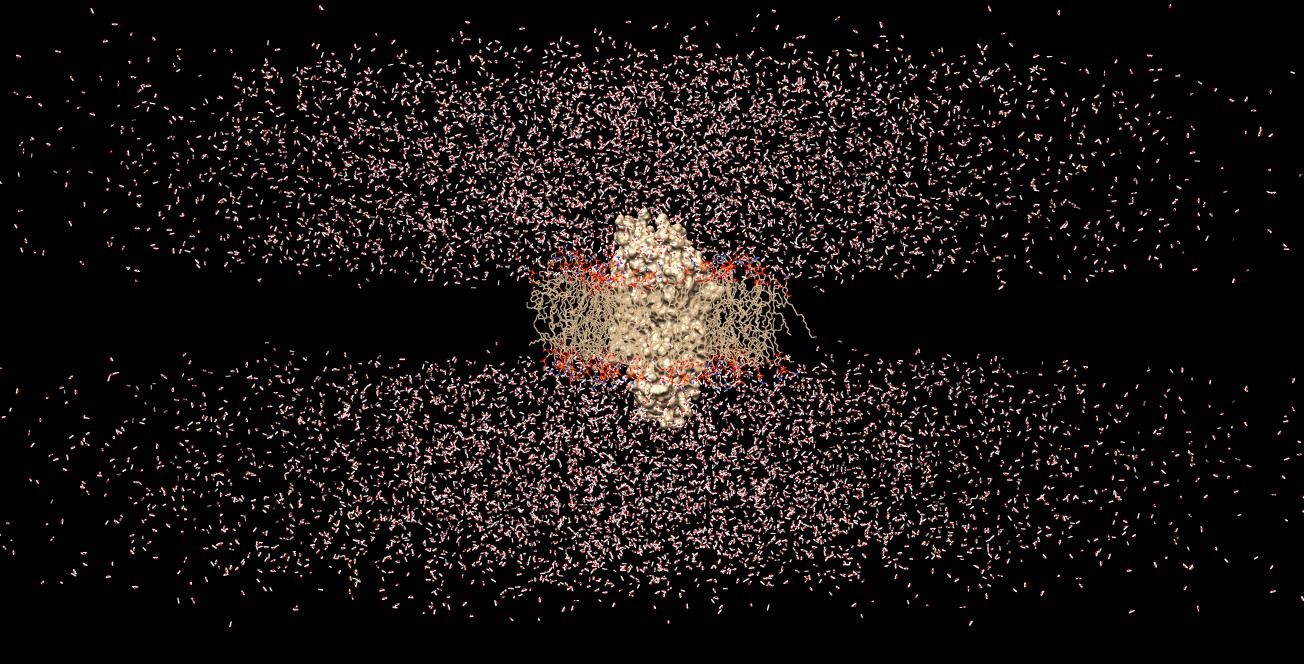
As a follow up to your suggestions, a 20 ns of production run (with all
restraints removed) was implemented using the lipid11 forces field for the
membrane part.
I attached a snapshot of the final step of the simulation.
Overall the production run went well (I guess!!), but there still seems
something weird.
Although the membrane and the protein+ligand complex seem to hold together,
this time the water layers above and below the membrane expanded
horizontally way too much, leaving the box ..
Is that expected for membrane simulations?
Is there a way to post-process water molecules in the trajectory so that
waters get back to the box.
Lastly, I tried parameterizing the membrane system using the lipid14
force field previously provided by Prof. Walker (Thanks again!). My
membrane system includes some cholesterol molecules. It appear that the
lipid14 force field does not include parameters for cholesterol. I was
wondering if you could also provide me with a cholesterol OFF file.
My best regards,
Gözde Yalçın
Res. Asst. in Drug Development Laboratory of Prof. Jenk Andac
2014-02-25 19:43 GMT+02:00 Gözde YALÇIN <yalcingozde88.gmail.com>:
> Dear Prof. Walker and Callum Dickson
> As a follow up to your suggestions, a 20 ns of production run (with all
> restraints removed) was implemented using the lipid11 forces field for the
> membrane part.
> I attached a snapshot of the final step of the simulation.
> Overall the production run went well (I guess!!), but there still seems
> something weird.
> Although the membrane and the protein+ligand complex seem to hold
> together, this time the water layers above and below the membrane expanded
> horizontally way too much, leaving the box ..
> Is that expected for membrane simulations?
> Is there a way to post-process water molecules in the trajectory so that
> waters get back to the box.
> Lastly, I tried parameterizing the membrane system using the lipid14
> force field previously provided by Prof. Walker (Thanks again!). My
> membrane system includes some cholesterol molecules. It appear that the
> lipid14 force field does not include parameters for cholesterol. I was
> wondering if you could also provide me with a cholesterol OFF file.
> My best regards,
> Gözde Yalçın
> Res. Asst. in Drug Development Laboratory of Prof. Jenk Andac
>
>
> 2014-02-12 19:42 GMT+02:00 Ross Walker <ross.rosswalker.co.uk>:
>
>> To follow up on what Callum says (I.e. your density equilibration is
>>
>> nowhere near long enough) - NPT simulations with lipid 11 need to be
>> NPgammaT with a constant surface tension term. See the example in the
>> tutorials section of the AMBER website.
>>
>> You may want to switch to Lipid14 - just released
>>
>> https://dl.dropboxusercontent.com/u/708185/lipid14_force_field_params_2014-
>> 01-30.tar.gz<https://dl.dropboxusercontent.com/u/708185/lipid14_force_field_params_2014-01-30.tar.gz>which should not require a constant surface tension term for
>> NPT. You should also probably be running the production run with
>> anisotropic NPT scaling unless you have specific reasons for wanting NVT.
>>
>> All the best
>> Ross
>>
>>
>>
>> On 2/12/14, 5:24 AM, "Gözde YALÇIN" <yalcingozde88.gmail.com> wrote:
>>
>> >Hi everyone,
>> >
>> >I did minimization and heating with these parameters and there is no
>> >problem at that part. However after 10 ps of pmemd.cuda run, it seems
>> that
>> >the lipids are falling apart as seen in the attached file. I was
>> wondering
>> >if I need to change parameters in cuda.in.
>> >
>> >relax.in
>> >Minimize the lipid structure
>> > &cntrl
>> > imin=1,
>> > maxcyc=10000,
>> > ncyc=5000,
>> > ntb=1,
>> > ntp=0,
>> > ntf=1,
>> > ntc=1,
>> > cut=10.0,
>> > ntpr=50,
>> > ntwr=2000,
>> > ioutfm=1,
>> > /
>> >temp1.in
>> >heating
>> > &cntrl
>> > imin=0, ntx=1, irest=0,
>> > ntc=2, ntf=2, tol=0.0000001,
>> > nstlim=2500, ntt=3, gamma_ln=1.0,
>> > ntr=1, ig=-1,
>> > ntpr=100, ntwr=10000, ntwx=100,
>> > dt=0.002, nmropt=1,
>> > ntb=1, ntp=0, cut=10.0, ioutfm=1,
>> > /
>> > &wt type='TEMP0', istep1=0, istep2=2500,
>> > value1=0.0, value2=100.0 /
>> > &wt type='END' /
>> >Hold lipid fixed
>> >10.0
>> >RES 231 581
>> >END
>> >END
>> >
>> >temp2.in
>> > heating
>> > &cntrl
>> > imin=0, ntx=5, irest=1,
>> > ntc=2, ntf=2,tol=0.0000001,
>> > nstlim=50000, ntt=3, gamma_ln=1.0,
>> > ntr=1, ig=-1,
>> > ntpr=100, ntwr=10000,ntwx=100,
>> > dt=0.002,nmropt=1,
>> > ntb=2,ntp=2,taup=2.0,cut=10.0,ioutfm=1,
>> > /
>> > &wt type='TEMP0', istep1=0, istep2=50000,
>> > value1=100.0, value2=300.0 /
>> > &wt type='END' /
>> >Hold lipid fixed
>> >10.0
>> >RES 231 581
>> >END
>> >END
>> >
>> >cuda.in
>> >cal Production MD NVE with
>> > GOOD energy conservation.
>> > &cntrl
>> > ntx=5, irest=1,
>> > ntc=2, ntf=2, tol=0.000001,
>> > nstlim=500000,
>> > ntpr=2500, ntwx=2500,
>> > ntwr=2500,
>> > dt=0.002, cut=8.,
>> > ntt=0, ntb=1, ntp=0,
>> > ioutfm=1,
>> > /
>> > &ewald
>> > dsum_tol=0.000001,
>> > /
>> >
>> >Can you help me to solve this problem.Thanks for your relevancy...
>> >
>> >Best regards...
>> >
>> >
>> >--
>> >Gözde YALÇIN
>> >Research Assistant
>> >Rize Recep Tayyip Erdogan University
>> >Faculty of Engineering
>> >Bioengineering Department
>> >53100 Rize-Turkey
>> >Ph.D. Student in Biotechnology
>> >Ankara University
>> >Biotechnology Institute
>> >06110 Besevler/Ankara-Turkey
>> >05065055074
>> >_______________________________________________
>> >AMBER mailing list
>> >AMBER.ambermd.org
>> >http://lists.ambermd.org/mailman/listinfo/amber
>>
>>
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
>>
>
>
>
> --
> Gözde YALÇIN
> Research Assistant
> Rize Recep Tayyip Erdogan University
> Faculty of Engineering
> Bioengineering Department
> 53100 Rize-Turkey
> Ph.D. Student in Biotechnology
> Ankara University
> Biotechnology Institute
> 06110 Besevler/Ankara-Turkey
> 05065055074
>
-- Gözde YALÇIN Research Assistant Rize Recep Tayyip Erdogan University Faculty of Engineering Bioengineering Department 53100 Rize-Turkey Ph.D. Student in Biotechnology Ankara University Biotechnology Institute 06110 Besevler/Ankara-Turkey 05065055074
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: 20ns-production-run.jpg)
