Date: Mon, 24 Feb 2014 04:37:02 +0000
Hi Everyone,
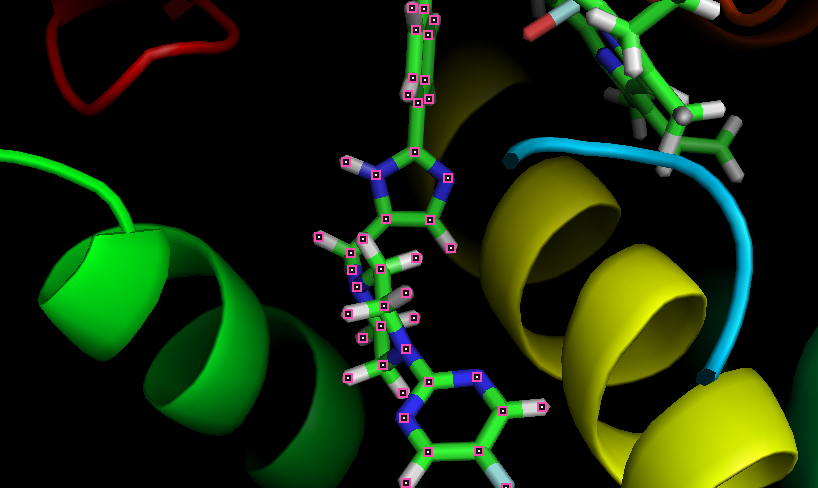
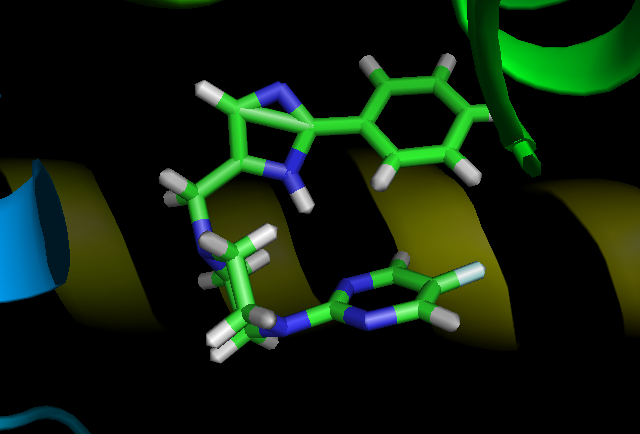
When visualizing my simulation in pymol I noticed that a new bond seems to form between two carbons in my ligand not present in the tleap output (paramaterized as cc and cd). Interestingly when I view the same pdb snapshot in UCSF Chimera or VMD the bond is not present.
While I don't believe this bond is real, as it isn't present in the original geometry optimized .mol2, params .frcmod or library fille, I'm wondering if anyone has experienced this before.
Attached to this email are snapshots of the tleap output (vis.png) as well as a point about 70 ns into my production run (prod.png). Additionally I've attached the .mol2 just in case I've missed something.
Any input would be greatly appreciated.
Best,
Parker
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: vis.png)
(image/png attachment: prod.png)
- application/octet-stream attachment: SCH.mol2
