Date: Tue, 1 Oct 2013 19:11:32 +0800
Dear amber users,
I am following the amber tutorial-
http://ambermd.org/tutorials/advanced/tutorial17/section1.htm for the
umbrella sampling of a dihedral angle of an alpha helical peptide from cis
to trans conformation.
The commands and the WHAM meta file are exactly same as given in this
tutorial. The only change is that, in the 1st simulation, dihedral angle of
starting structure is 0 degree (from crystal structure) and then we sampled
3 degree windows from 0 to 180 serially; while in 2nd simulation, dihedral
angle of starting structure is the 180 degree (structure extracted from 1st
simulation) and then we sampled 3 degree windows from 180 to 0 serially.
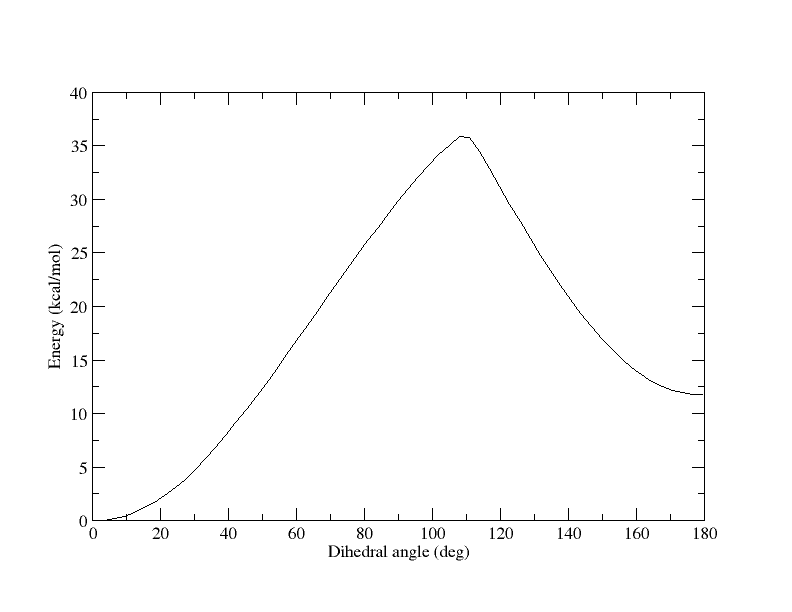
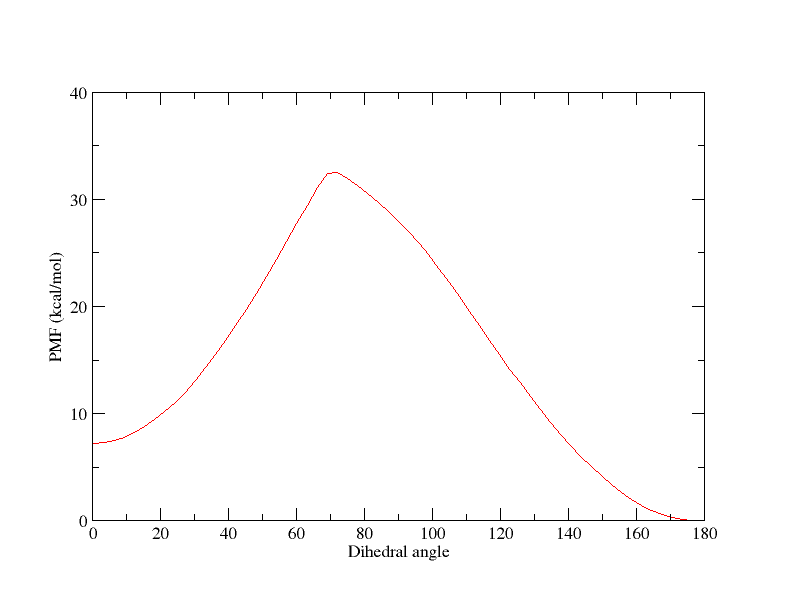
Each window is sampled for 500 ps in both the simulations. The PMF derived
from these calculations is quite contradictory to each other (plots
attached). Also, I ensure sufficient overlap of sampled dihedral angles
between consecutive windows.
Any suggestions why the PMF calculated from these 2 simulations is not
similar (even qualitatively)?
Thanks,
mohan
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: 0-180_pmf.png)
(image/png attachment: 180-0_pmf.png)
