Date: Thu, 19 Sep 2013 18:06:47 +0530
Dear Francois
I am also trying the commands you have mentioned earlier to me.
http://archive.ambermd.org/201209/0138.html
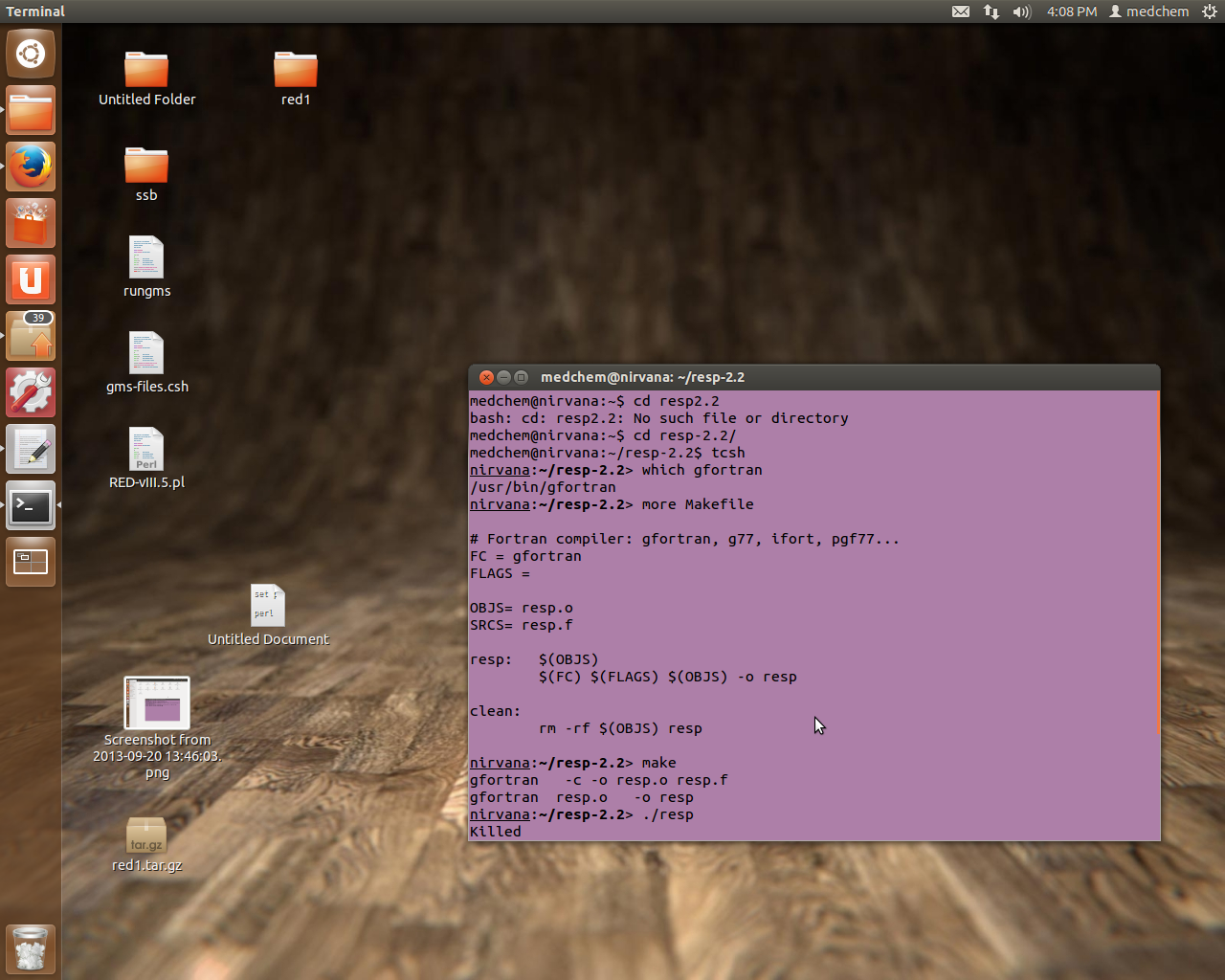
I am sending the snapshot
On Thu, Sep 19, 2013 at 5:54 PM, Shomesankar Bhunia <
rightclickatrighttime.gmail.com> wrote:
> Dear Francois,
> Thanks for the help. You are right. while typing resp in a terminal i am
> getting a reply "killed".
> while installing this i am doing this......
>
> cd resp-2.2
> make clean
> make
> su root
> cp resp /usr/local/bin/ (moving it to root)
> after this when I am typing "resp" I am getting a reply "killed"
> I am sending you my makefile.
> Than you for the support.
> Regards shome
>
>
> On Thu, Sep 19, 2013 at 5:42 PM, FyD <fyd.q4md-forcefieldtools.org> wrote:
>
>> Dear Shomesankar Bhunia,
>>
>> Thanks for your job/data...
>>
>> - the espot_m1 (MEP output to the RESP format) & the RESP inputs are
>> correctly generated
>> - the resp program is found (from the R.E.D. log file)
>>
>> but the RESP output are not generated: Where are the following output
>> files: output1_m1, output2_m1, punch1_m1 & punch2_m1
>>
>> It looks like the RESP program was not executed by R.E.D., or RESP was
>> not successfully compiled on your system...
>>
>> -1 which RESP version did you use? that available at
>> http://q4md-forcefieldtools.org/RED/resp/ or that from the AmberTools?
>>
>> -2 If I execute 'resp' in a terminal, I get:
>> lynx:~/red1> resp
>> usage: resp [-O] -i input -o output -p punch -q qin -t qout -e espot
>> -w qwts -s esout
>>
>> do you get a similar message?
>> (if not your RESP binary is not correctly compiled).
>>
>> regards, Francois
>>
>>
>>
>> > Did you use the last version of the R.E.D. III.x tools?
>> >
>> > Could you please send to my personal email address the entire R.E.D.
>> > III.x job (compressed archive file) with the Ante_R.E.D. & R.E.D. III.x
>> > source codes you used?
>> >
>> >
>> >> I am trying to derive RESP A1 charge for a ligand with Icharge=1. I
>> >> generated a GAMESS output (*.log)(generated by AntiRED) and used "
>> >> RED-vIII.5.pl" for calculation.
>> >> I got the following output.
>> >>
>> >> =================================================
>> >> ======================= Single molecule ===========================
>> >> The molecule TITLE is "MOLECULE"
>> >> The TOTAL CHARGE value of the molecule is "1"
>> >> The SPIN MULTIPLICITY value of the molecule is "1"
>> >>
>> >>
>> ===========================================================================
>> >>
>> >> * Selected optimization output *
>> >> GAMESS
>> >> Optimization OUTPUT looks nice !
>> >>
>> >> * 1 conformation(s) selected *
>> >>
>> >> WARNING:
>> >> A 2nd column of atom names is detected
>> >> This 2nd column will be used in the PDB (& Tripos) file(s)
>> >>
>> >> WARNING:
>> >> No three atom based re-orientation found in the P2N file
>> >> Re-orientation will be done according to the GAMESS Algorithm!
>> >>
>> >> * Selected QM Software *
>> >> GAMESS
>> >>
>> >> * Software checking *
>> >> gamess.00.x [ OK ]
>> >> rungms [ OK ]
>> >> ddikick.x [ OK ]
>> >> resp [ OK ]
>> >>
>> >> The Scratch directory defined for GAMESS is /tmp
>> >>
>> >> Scratch directory for GAMESS [ OK ]
>> >>
>> >>
>> >>
>> >> The punch file directory defined for GAMESS is $SCR/$JOB.dat
>> >>
>> >> MEP(s) is/are being computed for molecule 1 ... [ OK ]
>> >> See the file(s) "JOB2-gam_m1-1-(X).log"
>> >>
>> >> The RESP-A1 charges are being derived for molecule 1 ... [ FAILED
>> ]
>> >> See the "output(1|2)_m1" file(s)
>> >>
>> >>
>> >> Execution time: 0 h 50 m 3 s
>> >>
>> >> I looked at the output(1|2)_m1 file but i cant conclude anything. Its
>> like
>> >> this ( a part of file presented here)
>> >>
>> >> 51 2249 1 MOLECULE
>> >> 3.8981511E+00 -9.6413618E+00 -9.3606430E-01
>> >> 4.7466735E+00 -1.1461080E+01 -1.2320434E+00
>> >> 5.4224847E+00 -7.5141061E+00 -8.6283949E-01
>> >> 7.4220854E+00 -7.7083702E+00 -1.1036052E+00
>> >> 4.4816368E+00 -5.0735402E+00 -4.8522919E-01
>> >> 1.3078176E+00 -9.4270490E+00 -6.3449774E-01
>> >> 7.2522959E-02 -1.1035667E+01 -6.8510030E-01
>> >> 3.6637582E-01 -7.0338549E+00 -2.6151893E-01
>> >> 1.8661426E+00 -4.8828374E+00 -1.7454598E-01
>> >> 9.8373107E-02 -2.7055041E+00 2.7709774E-01
>> >> -2.3221161E+00 -3.7062019E+00 4.1801198E-01
>> >> -2.3179163E+00 -6.3330989E+00 9.4006139E-02
>> >> -4.1328556E+00 -7.6417417E+00 1.0079929E-01
>>
>>
>>
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
>>
>
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: 12.png)
