Date: Fri, 31 Aug 2012 19:47:14 +0200
Hi,
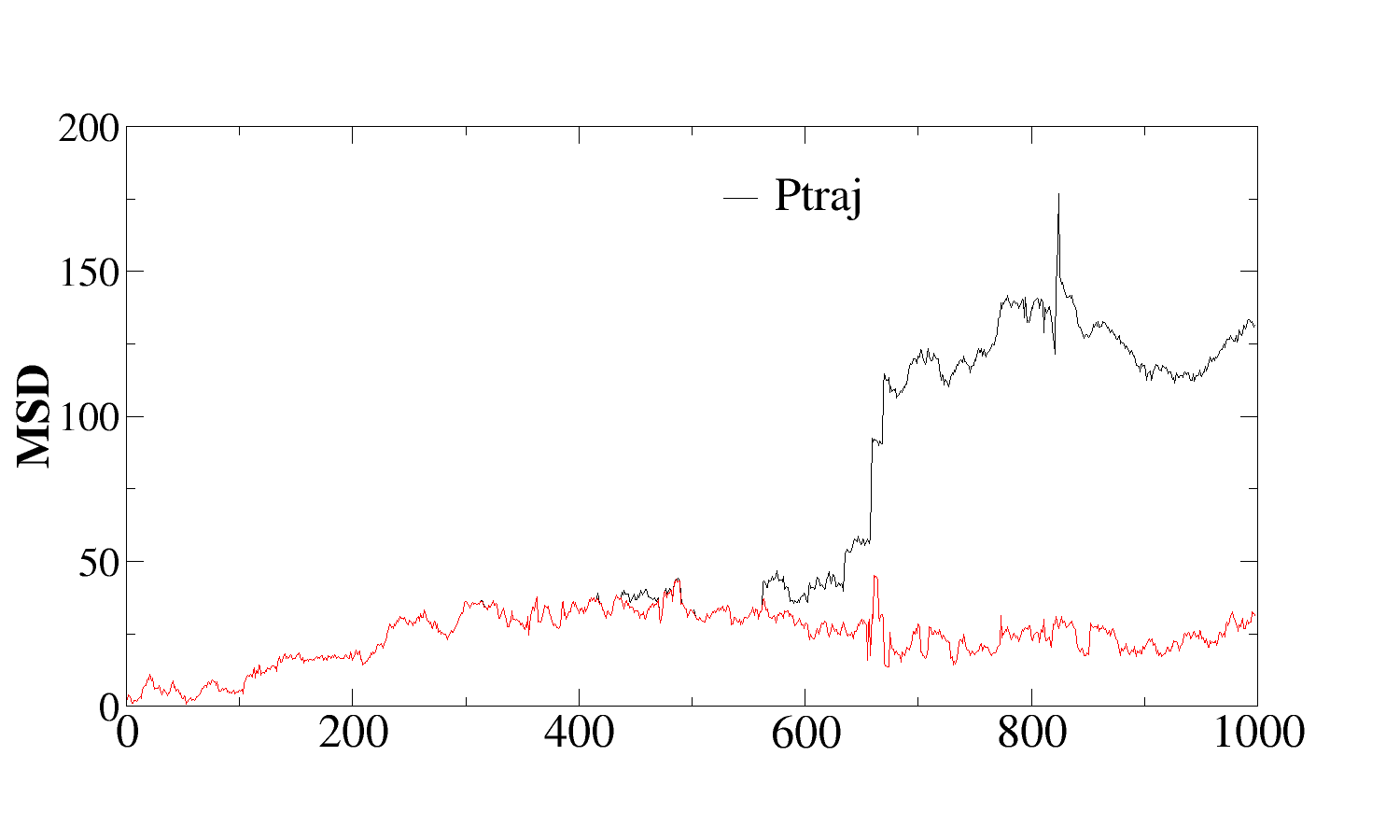
Thank you for the details. I have written small code to calculate msd
and compared with ptraj for a single water oxygen. I get the same
values upto a point and then a divergence (attached fig). I dnt know
if this could be due to periodicity problem.
I would like to know how periodicity is taken care of while
calculating the msd. Could someone please guide me on the same.
thanks,
Bala
On Sat, Aug 25, 2012 at 5:59 PM, Dmitry Mukha <dvmukha.gmail.com> wrote:
> Hi All,
>
> 2012/8/23 Daniel Roe <daniel.r.roe.gmail.com>:
>> Hi,
>>
>> >
>> More specifically, in the "<file>_a.xmgr" file the second column is
>> the average distance travelled by all the atoms, i.e. sqrt(
>> SUM[dist^2]/N ), where N is the number of atoms. Columns 3 and 4 in
>
> Actually, you are not right. sqrt(SUM[dist^2]/N )>=SUM[dist]/N, and a
> case of equality appears when 'dist'=const.
>
> In general form, for large numbers, mean square = square of mean + variance
>
> 2Bala:
>
> If you use 'average' key word, you will get only 2 columns, one for
> time and one for the appropriate mean value.
> _r - mean square displacement,
> _a - mean displacement,
> _x, _y, _z - Cartesian decomposition.
>
>> this case are the distance travelled by each individual atom, i.e.
>> sqrt( dist^2 ), so col2 should not be equal to the straight average of
>> cols 3 and 4, but rather col2 = sqrt( (col3^2 + col4^2) / 2), which
>> you can verify it is.
>>
>> Hope this helped.
>>
>> -Dan
>>
>> --
>
> --
> Sincerely,
> Dmitry Mukha
> Institute of Bioorganic Chemistry, NAS, Minsk, Belarus
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
-- C. Balasubramanian
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: MDA.png)
