Date: Fri, 11 Nov 2011 13:48:18 -0800
Dear all,
I am the beginner of Amber to do MD simulation for the ligand-receptor
pairs to examine the docking energy of this pair. My ligand is a small
molecule, and receptor is the riboswitch, a RNA molecule. I check the
crystal structure that indicates two Mg2+ are crucial to help binding
of ligand to the riboswitch, so I have to keep two Mg2+ ions for MD
simulation. In the pdb file, I keep the connections of these two Mg2+
ions to the RNA residues but to the ligand.
However, when I used xleap to add counterions, water box, and generate
prmtop and inpcrd files, xleap could not generate the parameter files
since there are the warning message (showed below). xleap seems like
not to recognize Mg2+ ions, and my ligand TPS.
I even tried to load gaff library, add counterions, water box, and
generate prmtop and inpcrd files, but xleap still could not make it,
with the same warning message. I do make sure that I loaded ions94.lib
to xleap. Another way to solve the problem, according to the old
conversation of archived message, is using ANTECHAMER to parameterize
the ligand and ions. But I always meet problems when parametering my
ligand in the beginning. The error message is this:
Running: /home/crystal/amber11/bin/bondtype -j full -i
ANTECHAMBER_BOND_TYPE.AC0 -o ANTECHAMBER_BOND_TYPE.AC -f ac
Info: Bond types are assigned for valence state 1 with penalty of 1
Running: /home/crystal/amber11/bin/atomtype -i ANTECHAMBER_AC.AC0 -o
ANTECHAMBER_AC.AC -p gaff
Total number of electrons: 179; net charge: 0
INFO: Number of electrons is odd: 179
Please check the total charge (-nc flag) and spin multiplicity (-m flag)
Running: /home/crystal/amber11/bin/sqm -O -i sqm.in -o sqm.out
Error: cannot run "/home/crystal/amber11/bin/sqm -O -i sqm.in -o
sqm.out" of bcc() in charge.c properly, exit
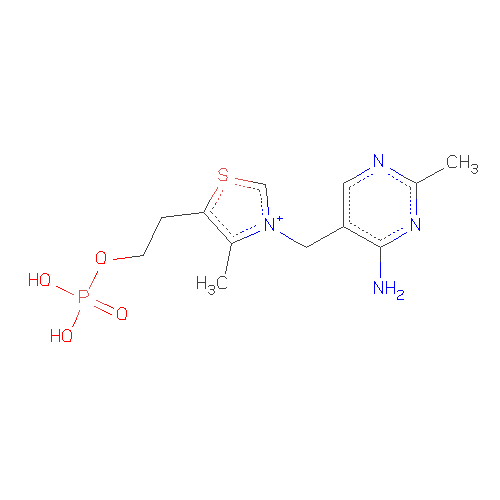
Antechamber seems not to precisely measure one positive charege on N
atom of the ligand TSP, based on the structure of the ligand. I have
no idea how to handle this problem. The structure is enclused in the
attachment.
I include the warning message after using the command "saveamberparm",
and the entire information from import pdb file, add counterions,
water box, following the warning message for your reference.
It is highly appreciated if someone could guide me to solve these
problems. I am glad of clarify something you feel confused because I
have met multiple problems of dealing with xleap and antechamber.
Sincerely,
Crystal
=====the warning message after use saveamberparm================
> saveamberparm 2hom 2HOM_MD_1.prmtop 2HOM_MD_1.inpcrd
Checking Unit.
WARNING: There is a bond of 8.309758 angstroms between:
------- .R<RU 25>.A<O3' 30> and .R<RG 26>.A<P 1>
WARNING: There is a bond of 3.013883 angstroms between:
------- .R<RG 75>.A<N1 20> and .R<MG 89>.A<MG 1>
FATAL: Atom .R<MG 89>.A<MG 1> does not have a type.
FATAL: Atom .R<MG 90>.A<MG 1> does not have a type.
FATAL: Atom .R<TPS 91>.A<CM2 1> does not have a type.
FATAL: Atom .R<TPS 91>.A<N3 2> does not have a type.
FATAL: Atom .R<TPS 91>.A<C2 3> does not have a type.
FATAL: Atom .R<TPS 91>.A<S1 4> does not have a type.
FATAL: Atom .R<TPS 91>.A<C5 5> does not have a type.
FATAL: Atom .R<TPS 91>.A<C4 6> does not have a type.
FATAL: Atom .R<TPS 91>.A<CM4 7> does not have a type.
FATAL: Atom .R<TPS 91>.A<C6 8> does not have a type.
FATAL: Atom .R<TPS 91>.A<C7 9> does not have a type.
FATAL: Atom .R<TPS 91>.A<O7 10> does not have a type.
FATAL: Atom .R<TPS 91>.A<N1A 11> does not have a type.
FATAL: Atom .R<TPS 91>.A<C2A 12> does not have a type.
FATAL: Atom .R<TPS 91>.A<N3A 13> does not have a type.
FATAL: Atom .R<TPS 91>.A<C4A 14> does not have a type.
FATAL: Atom .R<TPS 91>.A<N4A 15> does not have a type.
FATAL: Atom .R<TPS 91>.A<C5A 16> does not have a type.
FATAL: Atom .R<TPS 91>.A<C6A 17> does not have a type.
FATAL: Atom .R<TPS 91>.A<C7A 18> does not have a type.
FATAL: Atom .R<TPS 91>.A<P1 19> does not have a type.
FATAL: Atom .R<TPS 91>.A<O1 20> does not have a type.
FATAL: Atom .R<TPS 91>.A<O2 21> does not have a type.
FATAL: Atom .R<TPS 91>.A<O3 22> does not have a type.
Failed to generate parameters
Parameter file was not saved.
=====the entire information ===================================
Loading library: /home/crystal/amber11/dat/leap/lib/all_nucleic94.lib
Loading library: /home/crystal/amber11/dat/leap/lib/all_amino94.lib
Loading library: /home/crystal/amber11/dat/leap/lib/all_aminoct94.lib
Loading library: /home/crystal/amber11/dat/leap/lib/ions94.lib
Loading library: /home/crystal/amber11/dat/leap/lib/solvents.lib
> 2hom=loadpdb "2HOM_1_MD.pdb"
Loading PDB file: ./2HOM_1_MD.pdb
Enter zPdbReadScan from call depth 0.
Exit zPdbReadScan from call depth 0.
Matching PDB residue names to LEaP variables.
Mapped residue G, term: Terminal/beginning, seq. number: 0 to: RG5.
Mapped residue C, term: Nonterminal, seq. number: 1 to: RC.
Mapped residue G, term: Nonterminal, seq. number: 2 to: RG.
Mapped residue A, term: Nonterminal, seq. number: 3 to: RA.
Mapped residue C, term: Nonterminal, seq. number: 4 to: RC.
Mapped residue U, term: Nonterminal, seq. number: 5 to: RU.
Mapped residue C, term: Nonterminal, seq. number: 6 to: RC.
Mapped residue G, term: Nonterminal, seq. number: 7 to: RG.
Mapped residue G, term: Nonterminal, seq. number: 8 to: RG.
Mapped residue G, term: Nonterminal, seq. number: 9 to: RG.
Mapped residue G, term: Nonterminal, seq. number: 10 to: RG.
Mapped residue U, term: Nonterminal, seq. number: 11 to: RU.
Mapped residue G, term: Nonterminal, seq. number: 12 to: RG.
Mapped residue C, term: Nonterminal, seq. number: 13 to: RC.
Mapped residue C, term: Nonterminal, seq. number: 14 to: RC.
Mapped residue C, term: Nonterminal, seq. number: 15 to: RC.
Mapped residue U, term: Nonterminal, seq. number: 16 to: RU.
Mapped residue G, term: Nonterminal, seq. number: 17 to: RG.
Mapped residue C, term: Nonterminal, seq. number: 18 to: RC.
Mapped residue G, term: Nonterminal, seq. number: 19 to: RG.
Mapped residue U, term: Nonterminal, seq. number: 20 to: RU.
Mapped residue G, term: Nonterminal, seq. number: 21 to: RG.
Mapped residue A, term: Nonterminal, seq. number: 22 to: RA.
Mapped residue A, term: Nonterminal, seq. number: 23 to: RA.
Mapped residue G, term: Nonterminal, seq. number: 24 to: RG.
Mapped residue G, term: Nonterminal, seq. number: 25 to: RG.
Mapped residue C, term: Nonterminal, seq. number: 26 to: RC.
Mapped residue U, term: Nonterminal, seq. number: 27 to: RU.
Mapped residue G, term: Nonterminal, seq. number: 28 to: RG.
Mapped residue A, term: Nonterminal, seq. number: 29 to: RA.
Mapped residue G, term: Nonterminal, seq. number: 30 to: RG.
Mapped residue A, term: Nonterminal, seq. number: 31 to: RA.
Mapped residue A, term: Nonterminal, seq. number: 32 to: RA.
Mapped residue A, term: Nonterminal, seq. number: 33 to: RA.
Mapped residue U, term: Nonterminal, seq. number: 34 to: RU.
Mapped residue A, term: Nonterminal, seq. number: 35 to: RA.
Mapped residue C, term: Nonterminal, seq. number: 36 to: RC.
Mapped residue C, term: Nonterminal, seq. number: 37 to: RC.
Mapped residue C, term: Nonterminal, seq. number: 38 to: RC.
Mapped residue G, term: Nonterminal, seq. number: 39 to: RG.
Mapped residue U, term: Nonterminal, seq. number: 40 to: RU.
Mapped residue A, term: Nonterminal, seq. number: 41 to: RA.
Mapped residue U, term: Nonterminal, seq. number: 42 to: RU.
Mapped residue C, term: Nonterminal, seq. number: 43 to: RC.
Mapped residue A, term: Nonterminal, seq. number: 44 to: RA.
Mapped residue C, term: Nonterminal, seq. number: 45 to: RC.
Mapped residue C, term: Nonterminal, seq. number: 46 to: RC.
Mapped residue U, term: Nonterminal, seq. number: 47 to: RU.
Mapped residue G, term: Nonterminal, seq. number: 48 to: RG.
Mapped residue A, term: Nonterminal, seq. number: 49 to: RA.
Mapped residue U, term: Nonterminal, seq. number: 50 to: RU.
Mapped residue C, term: Nonterminal, seq. number: 51 to: RC.
Mapped residue U, term: Nonterminal, seq. number: 52 to: RU.
Mapped residue G, term: Nonterminal, seq. number: 53 to: RG.
Mapped residue G, term: Nonterminal, seq. number: 54 to: RG.
Mapped residue A, term: Nonterminal, seq. number: 55 to: RA.
Mapped residue U, term: Nonterminal, seq. number: 56 to: RU.
Mapped residue A, term: Nonterminal, seq. number: 57 to: RA.
Mapped residue A, term: Nonterminal, seq. number: 58 to: RA.
Mapped residue U, term: Nonterminal, seq. number: 59 to: RU.
Mapped residue G, term: Nonterminal, seq. number: 60 to: RG.
Mapped residue C, term: Nonterminal, seq. number: 61 to: RC.
Mapped residue C, term: Nonterminal, seq. number: 62 to: RC.
Mapped residue A, term: Nonterminal, seq. number: 63 to: RA.
Mapped residue G, term: Nonterminal, seq. number: 64 to: RG.
Mapped residue C, term: Nonterminal, seq. number: 65 to: RC.
Mapped residue G, term: Nonterminal, seq. number: 66 to: RG.
Mapped residue U, term: Nonterminal, seq. number: 67 to: RU.
Mapped residue A, term: Nonterminal, seq. number: 68 to: RA.
Mapped residue G, term: Nonterminal, seq. number: 69 to: RG.
Mapped residue G, term: Nonterminal, seq. number: 70 to: RG.
Mapped residue G, term: Nonterminal, seq. number: 71 to: RG.
Mapped residue A, term: Nonterminal, seq. number: 72 to: RA.
Mapped residue A, term: Nonterminal, seq. number: 73 to: RA.
Mapped residue G, term: Nonterminal, seq. number: 74 to: RG.
Mapped residue U, term: Nonterminal, seq. number: 75 to: RU.
Mapped residue C, term: Nonterminal, seq. number: 76 to: RC.
Mapped residue G, term: Nonterminal, seq. number: 77 to: RG.
Mapped residue C, term: Nonterminal, seq. number: 78 to: RC.
Mapped residue A, term: Terminal/last, seq. number: 79 to: RA3.
(Residue 80: MG, Terminal/beginning, was not found in name map.)
Unknown residue: MG number: 80 type: Terminal/beginning
..relaxing end constraints to try for a dbase match
-no luck
(Residue 81: MG, Nonterminal, was not found in name map.)
Unknown residue: MG number: 81 type: Nonterminal
(Residue 82: TPS, Terminal/last, was not found in name map.)
Unknown residue: TPS number: 82 type: Terminal/last
..relaxing end constraints to try for a dbase match
-no luck
Joining RG5 - RC
Joining RC - RG
Joining RG - RA
Joining RA - RC
Joining RC - RU
Joining RU - RC
Joining RC - RG
Joining RG - RG
Joining RG - RG
Joining RG - RG
Joining RG - RU
Joining RU - RG
Joining RG - RC
Joining RC - RC
Joining RC - RC
Joining RC - RU
Joining RU - RG
Joining RG - RC
Joining RC - RG
Joining RG - RU
Joining RU - RG
Joining RG - RA
Joining RA - RA
Joining RA - RG
Joining RG - RG
Joining RG - RC
Joining RC - RU
Joining RU - RG
Joining RG - RA
Joining RA - RG
Joining RG - RA
Joining RA - RA
Joining RA - RA
Joining RA - RU
Added missing heavy atom: .R<RU 43>.A<N1 13>
Added missing heavy atom: .R<RU 43>.A<C6 14>
Added missing heavy atom: .R<RU 43>.A<C2 22>
Added missing heavy atom: .R<RU 43>.A<C5 16>
Added missing heavy atom: .R<RU 43>.A<N3 20>
Added missing heavy atom: .R<RU 43>.A<O2 23>
Added missing heavy atom: .R<RU 43>.A<C4 18>
Added missing heavy atom: .R<RU 43>.A<O4 19>
Joining RU - RA
Joining RA - RC
Joining RC - RC
Joining RC - RC
Joining RC - RG
Joining RG - RU
Joining RU - RA
Joining RA - RU
Added missing heavy atom: .R<RU 51>.A<N1 13>
Added missing heavy atom: .R<RU 51>.A<C6 14>
Added missing heavy atom: .R<RU 51>.A<C2 22>
Added missing heavy atom: .R<RU 51>.A<C5 16>
Added missing heavy atom: .R<RU 51>.A<N3 20>
Added missing heavy atom: .R<RU 51>.A<O2 23>
Added missing heavy atom: .R<RU 51>.A<C4 18>
Added missing heavy atom: .R<RU 51>.A<O4 19>
Joining RU - RC
Added missing heavy atom: .R<RC 52>.A<N1 13>
Added missing heavy atom: .R<RC 52>.A<C6 14>
Added missing heavy atom: .R<RC 52>.A<C2 23>
Added missing heavy atom: .R<RC 52>.A<C5 16>
Added missing heavy atom: .R<RC 52>.A<N3 22>
Added missing heavy atom: .R<RC 52>.A<O2 24>
Added missing heavy atom: .R<RC 52>.A<C4 18>
Added missing heavy atom: .R<RC 52>.A<N4 19>
Joining RC - RA
Joining RA - RC
Joining RC - RC
Joining RC - RU
Joining RU - RG
Joining RG - RA
Joining RA - RU
Joining RU - RC
Joining RC - RU
Joining RU - RG
Joining RG - RG
Joining RG - RA
Joining RA - RU
Joining RU - RA
Joining RA - RA
Joining RA - RU
Joining RU - RG
Joining RG - RC
Joining RC - RC
Joining RC - RA
Joining RA - RG
Joining RG - RC
Joining RC - RG
Joining RG - RU
Added missing heavy atom: .R<RU 76>.A<N1 13>
Added missing heavy atom: .R<RU 76>.A<C6 14>
Added missing heavy atom: .R<RU 76>.A<C2 22>
Added missing heavy atom: .R<RU 76>.A<C5 16>
Added missing heavy atom: .R<RU 76>.A<N3 20>
Added missing heavy atom: .R<RU 76>.A<O2 23>
Added missing heavy atom: .R<RU 76>.A<C4 18>
Added missing heavy atom: .R<RU 76>.A<O4 19>
Joining RU - RA
Joining RA - RG
Joining RG - RG
Joining RG - RG
Joining RG - RA
Joining RA - RA
Joining RA - RG
Joining RG - RU
Joining RU - RC
Joining RC - RG
Joining RG - RC
Joining RC - RA3
Creating new UNIT for residue: MG sequence: 89
Created a new atom named: MG within residue: .R<MG 89>
Creating new UNIT for residue: MG sequence: 90
Starting new chain with
Created a new atom named: MG within residue: .R<MG 90>
Creating new UNIT for residue: TPS sequence: 91
Starting new chain with
Created a new atom named: CM2 within residue: .R<TPS 91>
Created a new atom named: N3 within residue: .R<TPS 91>
Created a new atom named: C2 within residue: .R<TPS 91>
Created a new atom named: S1 within residue: .R<TPS 91>
Created a new atom named: C5 within residue: .R<TPS 91>
Created a new atom named: C4 within residue: .R<TPS 91>
Created a new atom named: CM4 within residue: .R<TPS 91>
Created a new atom named: C6 within residue: .R<TPS 91>
Created a new atom named: C7 within residue: .R<TPS 91>
Created a new atom named: O7 within residue: .R<TPS 91>
Created a new atom named: N1A within residue: .R<TPS 91>
Created a new atom named: C2A within residue: .R<TPS 91>
Created a new atom named: N3A within residue: .R<TPS 91>
Created a new atom named: C4A within residue: .R<TPS 91>
Created a new atom named: N4A within residue: .R<TPS 91>
Created a new atom named: C5A within residue: .R<TPS 91>
Created a new atom named: C6A within residue: .R<TPS 91>
Created a new atom named: C7A within residue: .R<TPS 91>
Created a new atom named: P1 within residue: .R<TPS 91>
Created a new atom named: O1 within residue: .R<TPS 91>
Created a new atom named: O2 within residue: .R<TPS 91>
Created a new atom named: O3 within residue: .R<TPS 91>
total atoms in file: 1705
Leap added 899 missing atoms according to residue templates:
32 Heavy
867 H / lone pairs
The file contained 24 atoms not in residue templates
> addions 2hom Na+ 0
> solvateoct 2hom TIP3PBOX 10.0
> saveamberparm 2hom 2HOM_MD_1.prmtop 2HOM_MD_1.inpcrd
Checking Unit.
WARNING: There is a bond of 8.309758 angstroms between:
------- .R<RU 25>.A<O3' 30> and .R<RG 26>.A<P 1>
WARNING: There is a bond of 3.013883 angstroms between:
------- .R<RG 75>.A<N1 20> and .R<MG 89>.A<MG 1>
FATAL: Atom .R<MG 89>.A<MG 1> does not have a type.
FATAL: Atom .R<MG 90>.A<MG 1> does not have a type.
FATAL: Atom .R<TPS 91>.A<CM2 1> does not have a type.
FATAL: Atom .R<TPS 91>.A<N3 2> does not have a type.
FATAL: Atom .R<TPS 91>.A<C2 3> does not have a type.
FATAL: Atom .R<TPS 91>.A<S1 4> does not have a type.
FATAL: Atom .R<TPS 91>.A<C5 5> does not have a type.
FATAL: Atom .R<TPS 91>.A<C4 6> does not have a type.
FATAL: Atom .R<TPS 91>.A<CM4 7> does not have a type.
FATAL: Atom .R<TPS 91>.A<C6 8> does not have a type.
FATAL: Atom .R<TPS 91>.A<C7 9> does not have a type.
FATAL: Atom .R<TPS 91>.A<O7 10> does not have a type.
FATAL: Atom .R<TPS 91>.A<N1A 11> does not have a type.
FATAL: Atom .R<TPS 91>.A<C2A 12> does not have a type.
FATAL: Atom .R<TPS 91>.A<N3A 13> does not have a type.
FATAL: Atom .R<TPS 91>.A<C4A 14> does not have a type.
FATAL: Atom .R<TPS 91>.A<N4A 15> does not have a type.
FATAL: Atom .R<TPS 91>.A<C5A 16> does not have a type.
FATAL: Atom .R<TPS 91>.A<C6A 17> does not have a type.
FATAL: Atom .R<TPS 91>.A<C7A 18> does not have a type.
FATAL: Atom .R<TPS 91>.A<P1 19> does not have a type.
FATAL: Atom .R<TPS 91>.A<O1 20> does not have a type.
FATAL: Atom .R<TPS 91>.A<O2 21> does not have a type.
FATAL: Atom .R<TPS 91>.A<O3 22> does not have a type.
Failed to generate parameters
Parameter file was not saved.
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/gif attachment: TPS_500.gif)
