Date: Thu, 24 Mar 2011 14:50:08 +0800
Hello,
I have two questions to consult.
Firstly, I want to do the molecular dynamic simulation between RNA and its ligand, so I uploaded the complex of the RNA and the ligand. Using leaprc.ff03 force field, there is no corresponding parameters for U. How can I do? Choose another force filed, such as leaprc.rna.ff02?
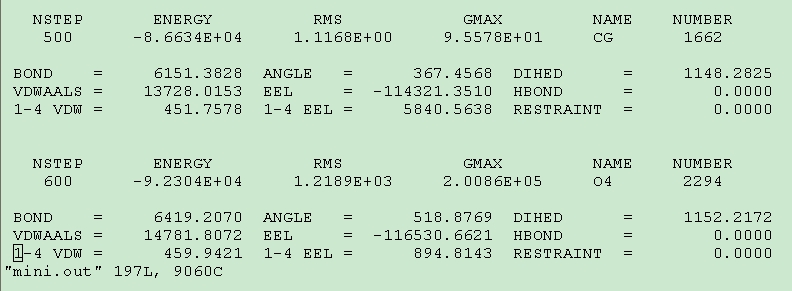
Secondly, I do the minimization of some complexes. Some of them have abnormal termination without errors at the bottom of the output file. The following are the input file and the part of output file. How can I handle this problem?
Thank you very much!
Best wishes!
mini.in
mini.out
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber

(image/jpeg attachment: Catch2.jpg)
(image/jpeg attachment: Catch1_03-24-12-04-36_.jpg)
