Date: Thu, 18 May 2006 10:33:35 -0700
Dear AMBER users,
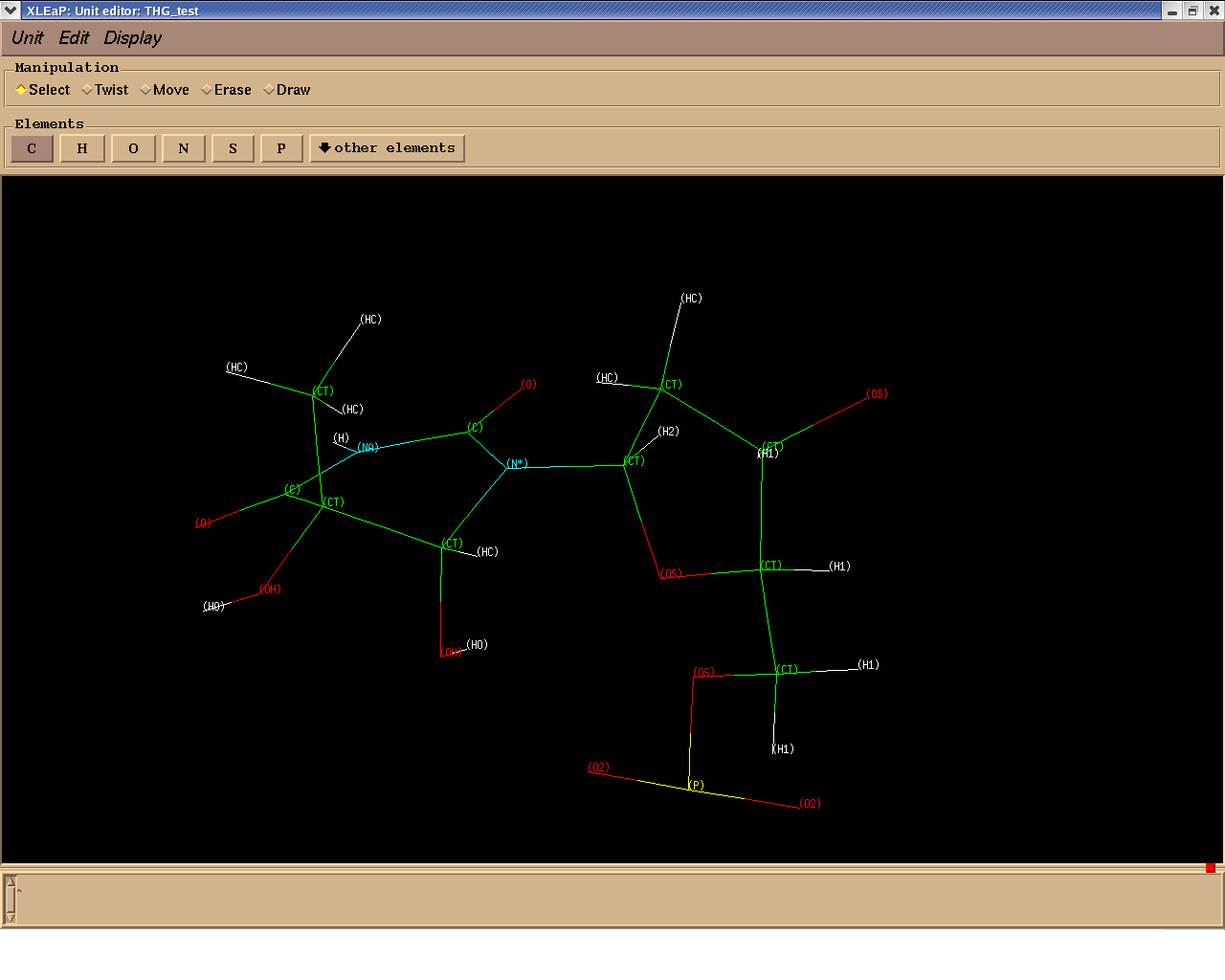
I am running an MD simulation for a non standard nucleic acid which is
non planar.
In this structure, I have a methyl group in a pseudo axial position.
My question is:
1) how can I prevent this methyl from inverting to the equatorial
position.
The improper dihedrals I used for these simulations are as follow:
CT-C-N*-CT 1.1 180.0 2.0
CT-C-N*-CM 1.1 180.0 2.0
C-CT-CT-CT 1.1 180.0 2.0
N*-OH-CT-HC 1.1 180.0 2.0
CT-OH-CT-HC 1.1 180.0 2.0
N*-CT-CT-HC 1.1 180.0 2.0
2) May the proper dihedral angles also play a role in this transition
Attached is the structure of this system with the atom types assigned. I
might be doing something wrong,
any advice will be welcome
Latifa
-----------------------------------------------------------------------
The AMBER Mail Reflector
To post, send mail to amber.scripps.edu
To unsubscribe, send "unsubscribe amber" to majordomo.scripps.edu
(image/png attachment: thg_types.png)
